CP2K
Description¶
CP2K is a quantum chemistry and solid state physics software package that can perform atomistic simulations of solid state, liquid, molecular, periodic, material, crystal, and biological systems. CP2K provides a general framework for different modeling methods such as DFT using the mixed Gaussian and plane waves approaches GPW and GAPW.
Versions¶
Following versions of CP2K are currently available:
- Runtime dependencies:
You can load the CP2K module by following command:
module load CP2K/2022.1-foss-2022a
User guide¶
To know more information about CP2K tutorial and documentation, please refer to CP2K HOWTOs.
Benchmarks¶
CP2K's grid-based calculation as well as DBCSR's block sparse matrix multiplication (Cannon algorithm) prefer a square-number for the total rank-count (2d communication pattern).
It can be more efficient to leave CPU-cores unused in order to achieve this square-number property rather than using all cores with a "non-preferred" total rank-count Counter-intuitively, even an unbalanced rank-count per node i.e., different rank-counts per socket can be an advantage. Pinning MPI processes and placing threads requires extra care to be taken on a per-node basis to load a dual-socket system in a balanced fashion or to setup space between ranks for the OpenMP threads.
In order to better understand how CP2K utilises the available hardware on Devana and how to get good performance we can examine the effect on benchmark performance of the choice of the number of MPI ranks per node and OpenMP thread.
Following command has been used to run the benchmarks:
mpiexec -np $mpi_rank --map-by slot:PE=$omp_thread -x OMP_PLACES=cores -x OMP_PROC_BIND=SPREAD -x OMP_NUM_THREADS=$omp_thread cp2k.psmp -i ${benchmark}.inp > ${benchmark}.out
Working directory
Single-node benchmarks have been run on local /work/ storage native to each compute node, which are generally faster than shared storage hosting /home/ and /scratch/ directories.
Benchmarks have been made on following systems:
Ab-initio molecular dynamics of liquid water using the Born-Oppenheimer approach, using Quickstep DFT. Production quality settings for the basis sets (TZV2P) and the planewave cutoff (280 Ry) are chosen, and the Local Density Approximation (LDA) is used for the calculation of the Exchange-Correlation energy. The configurations were generated by classical equilibration, and the initial guess of the electronic density is made based on Atomic Orbitals. The single-node system contains 64 water molecules (192 atoms, 512 electrons) in a 12.4 Å3 cell and the cross-node system contains 526 water molecules (768 atoms, 2048 electrons) in 19.7 Å3 cell. Both MDs are run for 10 steps.
| Single node Performance | Cross-node Performance |
|---|---|
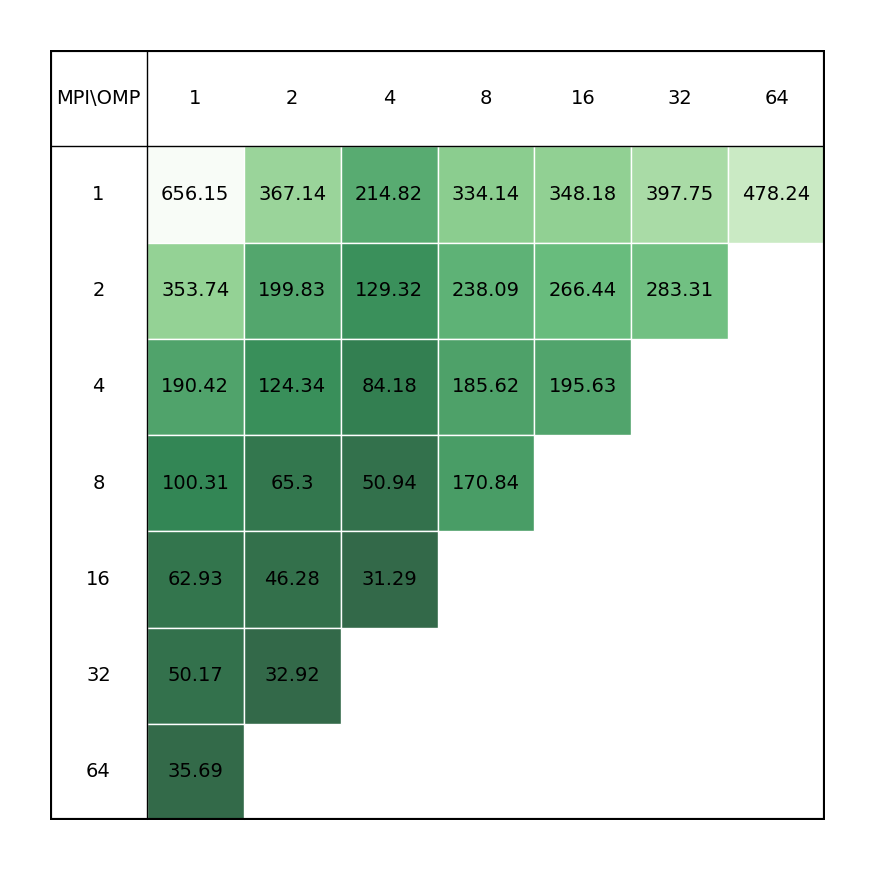 |
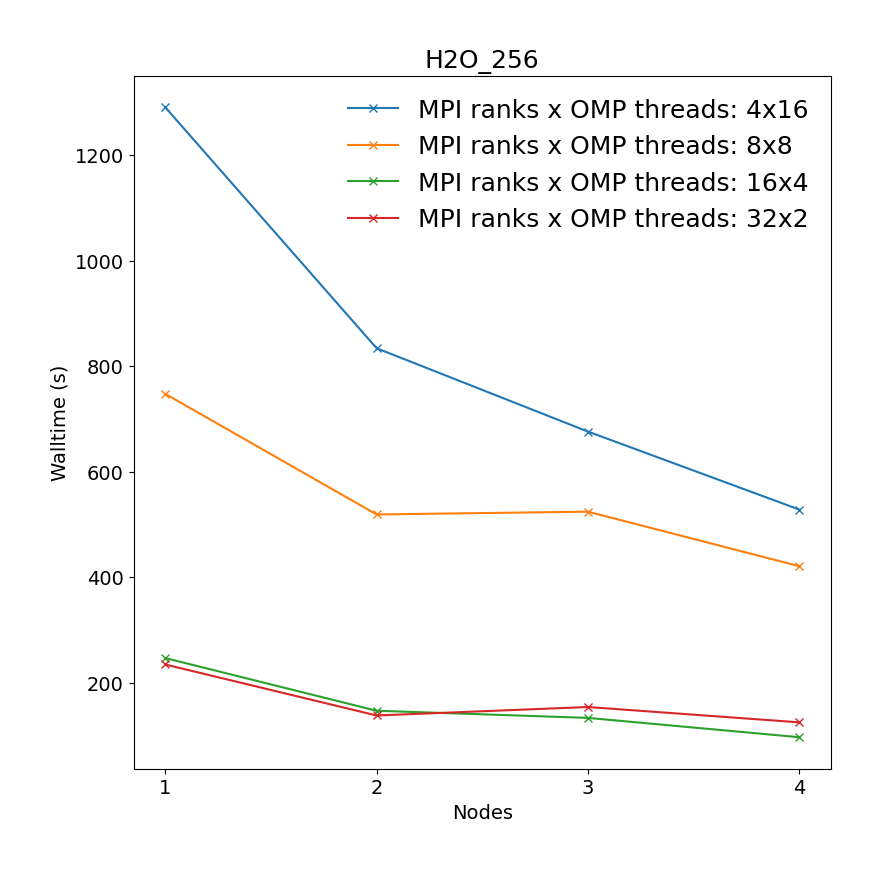 |
Note
"Change of the system for cross-node benchmarks was done due to relatively small system selected for single-node calculations."
This is a short molecular dynamics run of 1000 time steps in a NPT ensemble at 300K. It consists of 28000 atoms - a 103 supercell with 28 atoms of iron silicate (Fe2SiO4, also known as Fayalite) per unit cell. The simulation employs a classical potential (Morse with a hard-core repulsive term and 5.5 Å cutoff) with long-range electrostatics using Smoothed Particle Mesh Ewald (SPME) summation. While CP2K does support classical potentials via the Frontiers In Simulation Technology (FIST) module, this is not a typical calculation for CP2K but is included to give an impression of the performance difference between machines for the MM part of a QM/MM calculation.
| Single node Performance | Cross-node Performance |
|---|---|
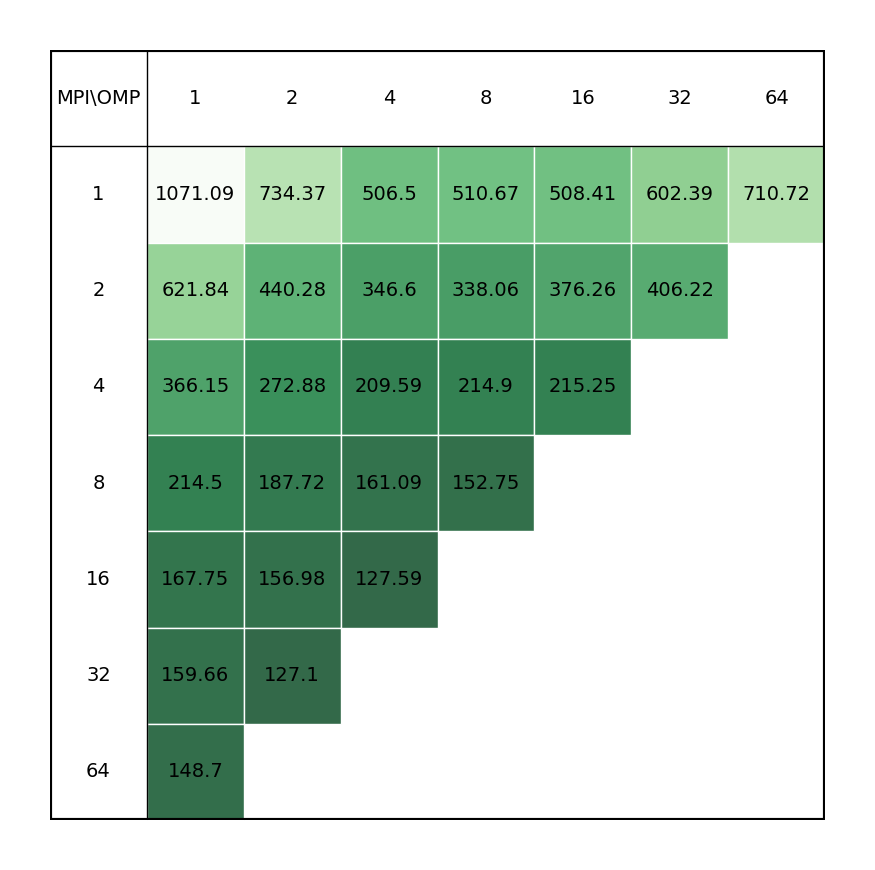 |
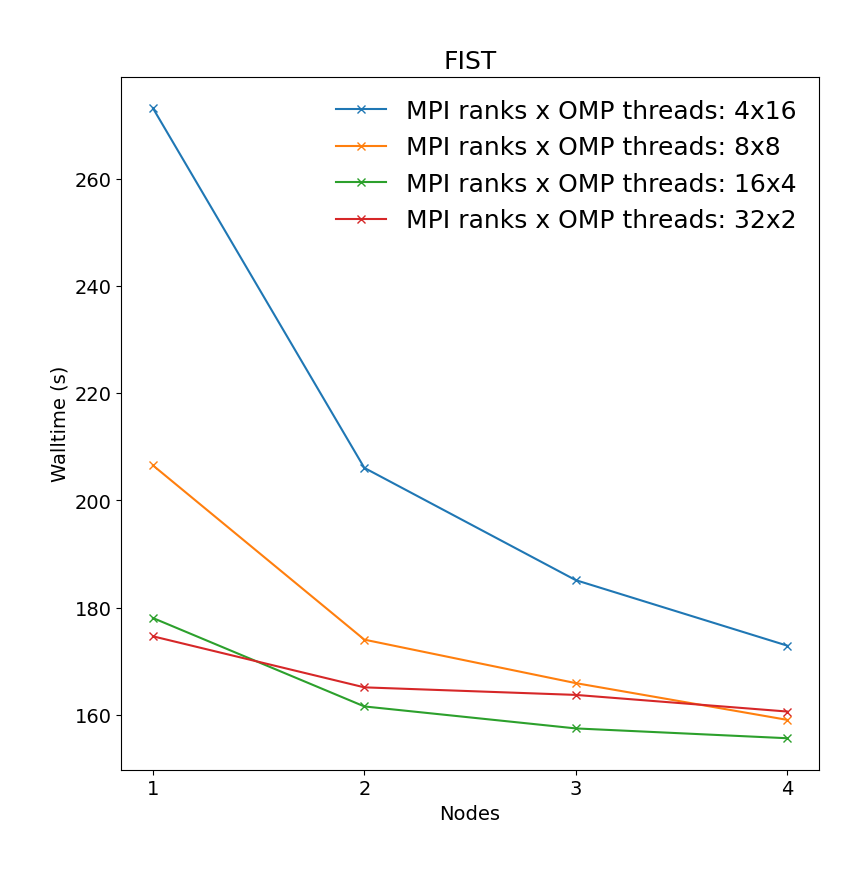 |
Note
"Performance drop from single node performance table on left and cross-node performance measured for one node is caused by different disks used during calculations, i.e., single node performance was calculated using local /work/ disk, while cross-node performance was calculated on shared /scratch/ storage."
This is a single-point energy calculation using linear-scaling DFT. It consists of 6144 atoms in a 39 Å3 box (2048 water molecules in total). An LDA functional is used with a DZVP MOLOPT basis set and a 300 Ry cut-off. For large systems the linear-scaling approach for solving Self-Consistent-Field equations will be much cheaper computationally than using standard DFT and allows scaling up to 1 million atoms for simple systems. The linear scaling cost results from the fact that the algorithm is based on an iteration on the density matrix. The cubically-scaling orthogonalisation step of standard Quickstep DFT using OT is avoided and the key operation is sparse matrix-matrix multiplications, which have a number of non-zero entries that scale linearly with system size. These are implemented efficiently in the DBCSR library.
| Single node Performance | Cross-node Performance |
|---|---|
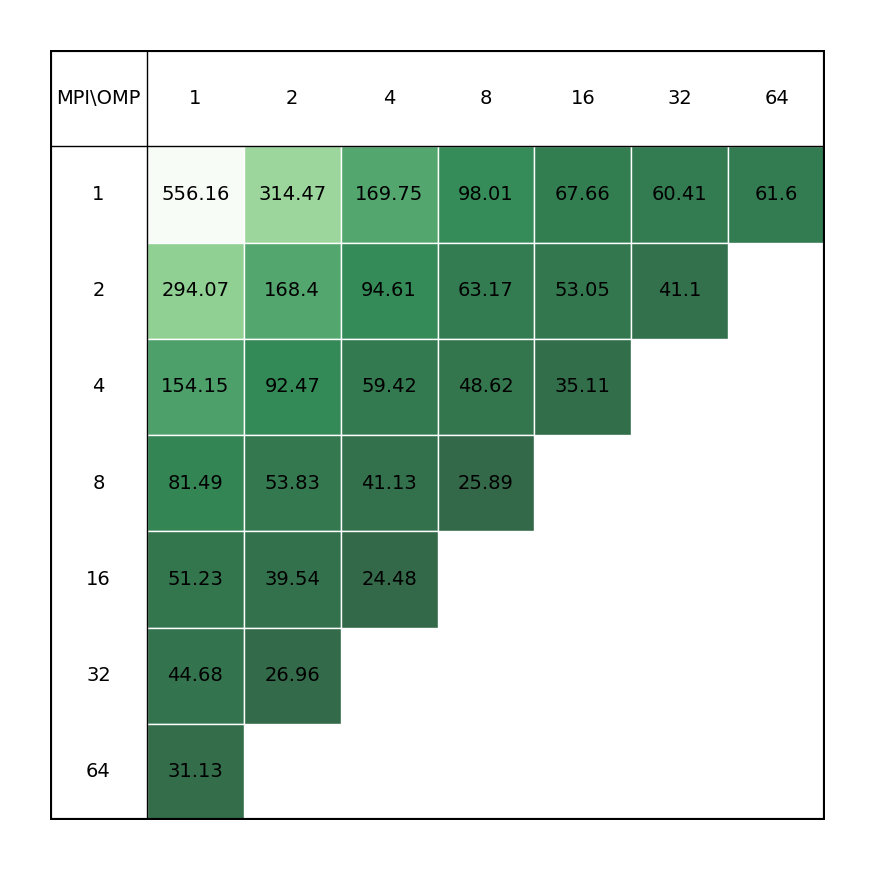 |
 |
Because of the above-mentioned complexity, a script for planning MPI/OpenMP-hybrid execution CP2K_plan.sh is made available to users located in /storage-apps/scripts/. For a system with 64 cores per node (two sockets, disabled hyperthreading), setting up the "system metric" looks like:
./CP2K_plan.sh <num-nodes> <ncores-per-node> <nthreads-per-core> <nsockets-per-node>:
./CP2K_plan.sh 1 64 1 2
./CP2K_plan.sh 2 128 1 2
Example run script¶
You can copy and modify this script to cp2k_run.sh and submit job to a compute node by command sbatch cp2k_run.sh.
#!/bin/bash
#SBATCH -J "CP2K" # name of job in SLURM
#SBATCH --account=<project> # project number
#SBATCH --partition= # selected partition (short, medium, long)
#SBATCH --nodes= # number of nodes
#SBATCH --ntasks= # number of mpi ranks, needs to be tested for the best performance
#SBATCH --cpus-per-task= # number of cpus per mpi rank, needs to be tested for the best performance
#SBATCH --time=hh:mm:ss # time limit for a job
#SBATCH -o stdout.%J.out # standard output
#SBATCH -e stderr.%J.out # error output
# Modules
module load CP2K/2022.1-foss-2022a
srun -n ${SLURM_NTASKS} cp2k.popt -i example.inp > outputfile.out
mpiexec -np ${SLURM_NTASKS} --map-by slot:PE=${SLURM_CPUS_PER_TASK} -x OMP_PLACES=cores -x OMP_PROC_BIND=SPREAD -x OMP_NUM_THREADS=${SLURM_CPUS_PER_TASK} cp2k.psmp -i ${benchmark}.inp > ${benchmark}.out
GPU accelerated CP2K¶
Note
" Section under construction"