GROMACS
Description¶
GROMACS is a versatile package to perform molecular dynamics for systems with hundreds to millions of particles. It is primarily designed for biochemical molecules like proteins, lipids and nucleic acids that have a lot of complicated bonded interactions, but since GROMACS is extremely fast at calculating the nonbonded interactions it can also be used for dynamics of non-biological systems, such as polymers and fluid dynamics.
Versions¶
Following versions of GROMACS are currently available:
- Runtime dependencies:
You can load selected version (also with runtime dependencies) as a one module by following commands:
module load GROMACS/2021.5-foss-2021b
- Runtime dependencies:
You can load selected version (also with runtime dependencies) as a one module by following commands:
module load GROMACS/2023.2-intelmkl-CUDA-12.0
Info
Version 2021.5-foss-2021b is deprecated build without support for efficient thread-MPI parallelization thread-MPI and users are strongly encouraged to use newer builds with enable MPI support.
User guide¶
You can find the software documentation and user guide on the official gromacs website.
Additionally, excellent GROMACS tutorials for new, as well as advanced users, can be accessed on this link.
Finally, the most comprehensive up-to-date documentation, containing building options, user guides, keywords explanation, and error debugging, and can be found on the following page.
Benchmarks¶
In order to better understand how GROMACS utilises the available hardware on Devana and how to get good performance we can examine the effect on benchmark performance of the choice of the number of MPI ranks per node and OpenMP thread.
Following command has been used to run the benchmarks:
mpiexec -np $ntmpi gmx_mpi mdrun -v -s $trajectory.tpr -ntomp $ntomp -pin on -nsteps 20000 -deffnm $trajectory
For more information about these benchmarks systems see following page.
Info
"Single-node benchmarks have been run on local /work/ storage native to each compute node, which are generally faster than shared storage hosting /home/ and /scratch/ directories."
Benchmarks have been made on following systems:
Molecular dynamics simulation of protein in membrane surrounded by water molecules (81743 atoms with system size 10.8 x 10.2 x 9.6 Å3) with a 2fs time step for a total of 40ps. All bonds constraints were utilized during benchmark, i.e.m the update step had to be done on CPU.
| Single node Performance | Cross-node Performance |
|---|---|
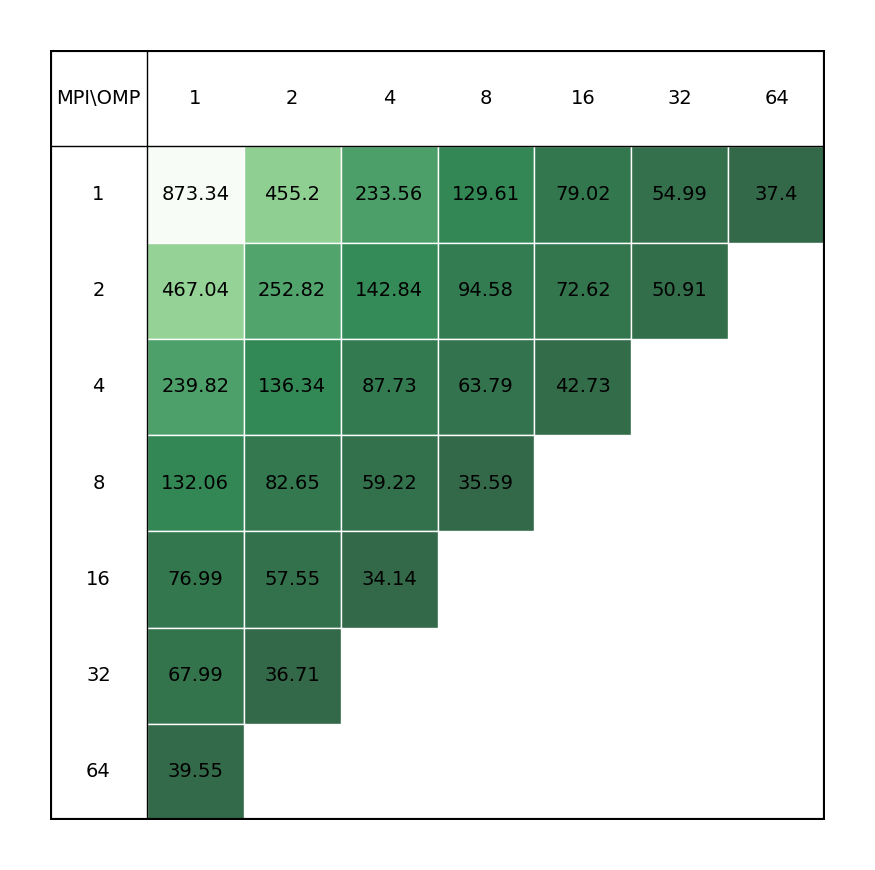 |
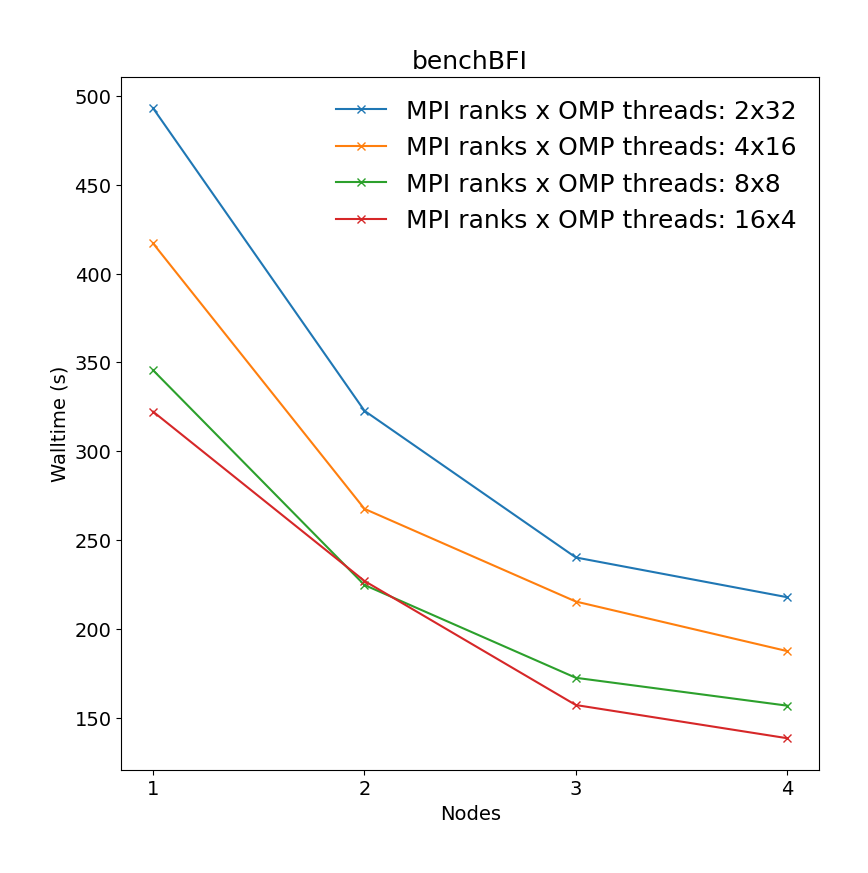 |
Binding affinity study benchmark of protein-ligand system surrounded by water molecules (ca. 107k atoms) with energy evaluations done every step (TI). All bonds constraints were utilized during benchmark, i.e.m the update step had to be done on CPU.
| Single node Performance | Cross-node Performance |
|---|---|
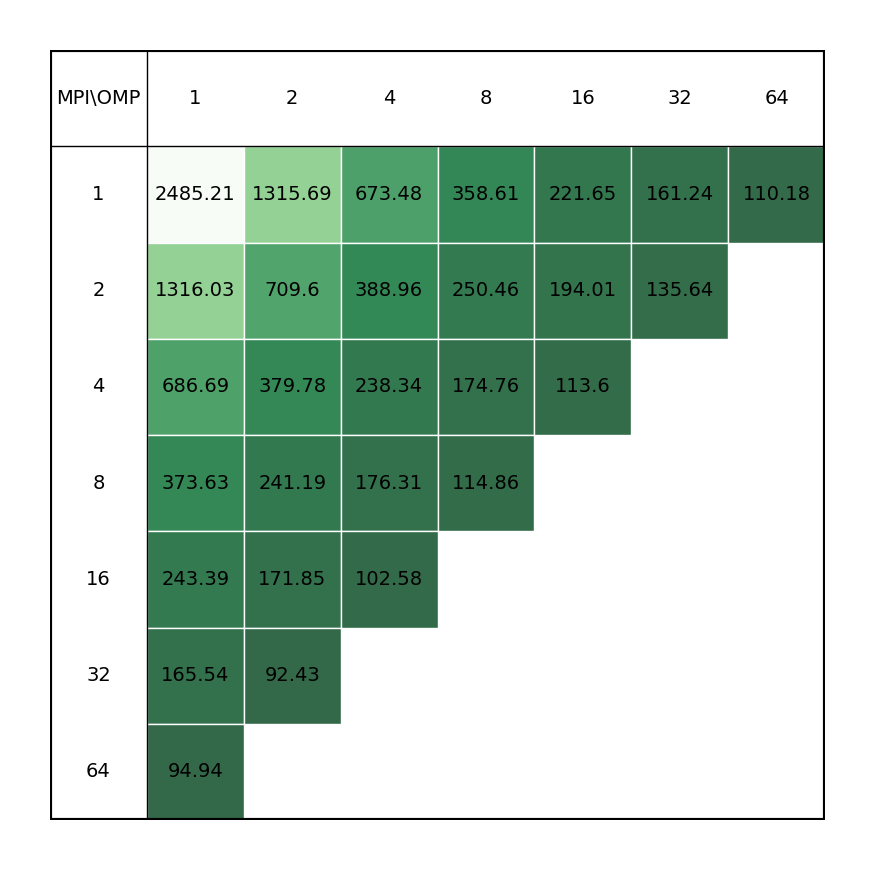 |
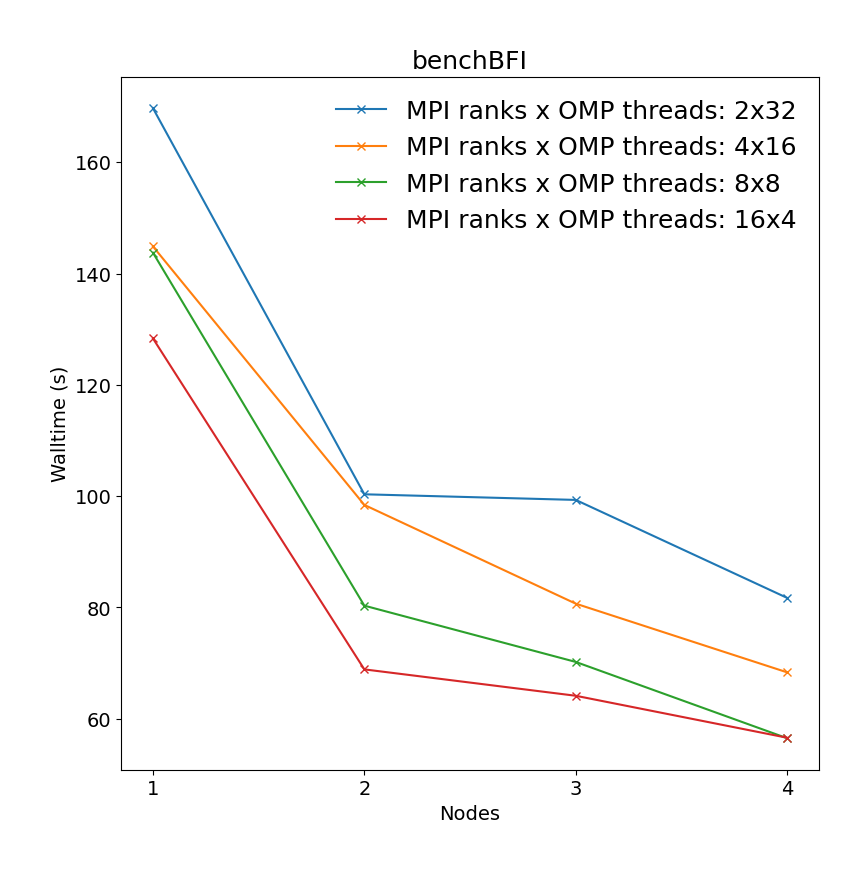 |
Binding affinity study benchmark of bromosporine to bromodomain surrounded by water molecules (43952 atoms with system size 8.55 x 8.55 x 6.04 Å3) with a 2fs time step for a total of XXps. Free energy is controlled with init-lambda-state, coul-lambdas and vdw-lambdas vectors, all 20 lambda neighbors are calculated, energy evaluations done every step. All bonds constraints were utilized during benchmark, i.e.m the update step had to be done on CPU.
| Single node Performance | Cross-node Performance |
|---|---|
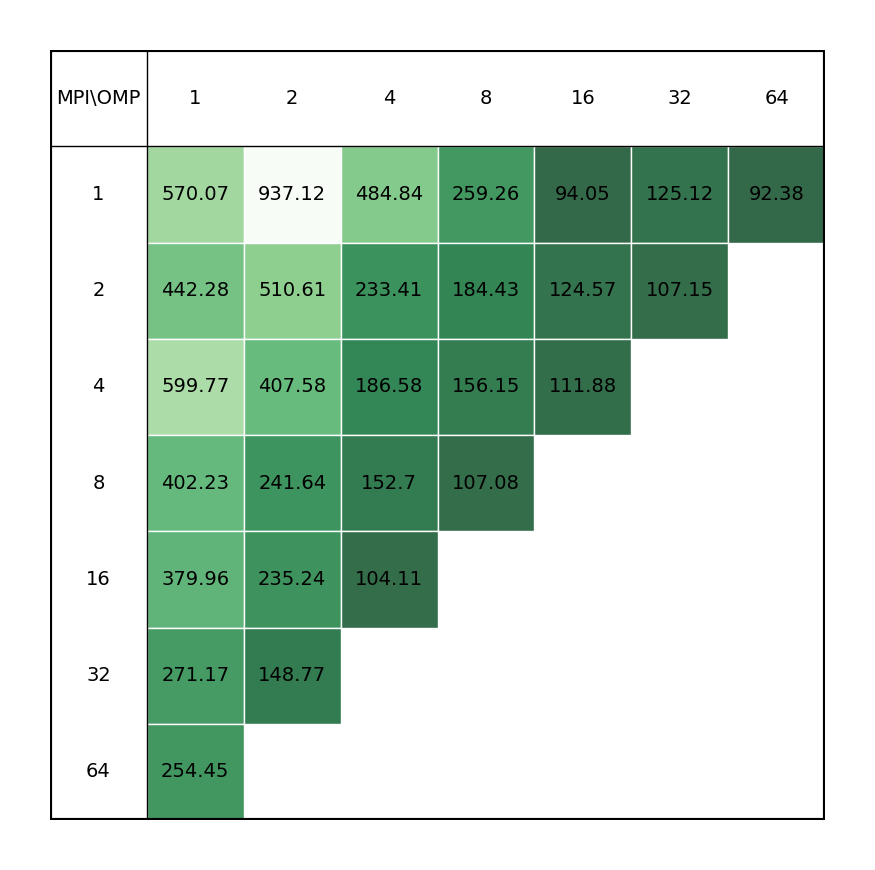 |
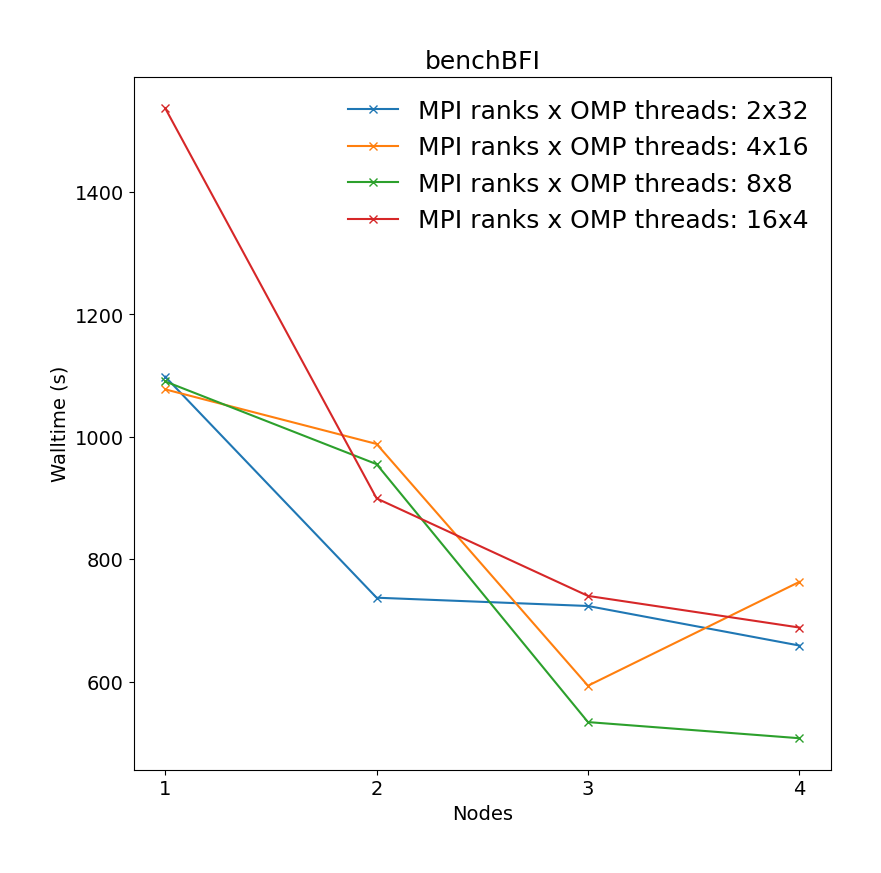 |
It is clear there is a very significant effect on performance of the choice of MPI x OpenMP hybrid decomposition. It is important to understand that diagonals correspond to total BUs used, i.e. the outermost diagonal corresponds to 64 BUs, the second outermost to 32 BUs, etc. As a general rule performance on Devana cluster is best achieved with 16 OpenMP threads per MPI ranks. Hence, the user can control alloacted portion of node by selecting number of MPI ranks from 1 to 4.
Example run script¶
You can copy and modify this script to gromacs_run.sh and submit job to a compute node by command sbatch gromacs_run.sh.
#!/bin/bash
#SBATCH -J "gromacs_job" # name of job in SLURM
#SBATCH --account=<project> # project number
#SBATCH --partition= # selected partition (short, medium, long)
#SBATCH --nodes= # number of nodes
#SBATCH --ntasks= # number of mpi ranks, needs to be tested for the best performance
#SBATCH --cpus-per-task= # number of cpus per mpi rank, needs to be tested for the best performance
#SBATCH --time=hh:mm:ss # time limit for a job
#SBATCH -o stdout.%J.out # standard output
#SBATCH -e stderr.%J.out # error output
module load GROMACS/2023.2-intelmkl-CUDA-12.0
# Modify according to specific needs
init_dir=`pwd`
work_dir=/scratch/$USER/GROMACS/$SLURM_JOB_ID
# Copy files over
input_files=""
output_files=""
# Move to working directory
cd $work_dir
cp $input_files $work_dir/.
# Start GROMACS
mpiexec -np ${SLURM_NTASKS} gmx_mpi mdrun -ntomp ${SLURM_CPUS_PER_TASK} ..
# Move files back
cp $output_files $init_dir/.
GPU accelerated GROMACS¶
GPU support has been implemented in GROMACS since version 5.* and version GROMACS/2023.2-intelmkl-CUDA-12.0 has been compiled with CUDA/GPU support.
Benchmarks¶
Note
"Section under construction."
For more information about GPU benchmarks systems see following page.
Example run script¶
#!/bin/bash
#SBATCH -J "gromacs_gpu_job" # name of job in SLURM
#SBATCH --account=<project> # project number
#SBATCH --partition=ngpu # select gpu nodes
#SBATCH --nodes= # number of nodes
#SBATCH --ntasks= # number of mpi ranks, needs to be tested for the best performance
#SBATCH --cpus-per-task= # number of cpus per mpi rank, needs to be tested for the best performance
#SBATCH --time=hh:mm:ss # time limit for a job
#SBATCH -o stdout.%J.out # standard output
#SBATCH -e stderr.%J.out # error output
module load GROMACS/2023.2-intelmkl-CUDA-12.0
# Modify according to specific needs
init_dir=`pwd`
work_dir=/scratch/$USER/GROMACS/$SLURM_JOB_ID
# Copy files over
input_files=""
output_files=""
# Move to working directory
cd $work_dir
cp $input_files $work_dir/.
# Start GROMACS
mpiexec -np ${SLURM_NTASKS} gmx_mpi mdrun -ntomp ${SLURM_CPUS_PER_TASK} -nb gpu -pme gpu ..
# Move files back
cp $output_files $init_dir/.
The script offloads computation of short range nonbonded interactions to GPU (-nb gpu), which provides the majority of the available speed-up compared to run using only the CPU. Secondly offloading of the PME calculation to the GPU (-pme gpu), serves to further reduce the load on the CPU and improve usage overlap between CPU and GPU. Bonded interactions can also be assigend to GPU **(-bonded gpu), when the amount of CPU resources per GPU is relatively little (either because the CPU is weak or there are few CPU cores assigned to a GPU in a run). See GROMACS manual "Running mdrun with GPUs" for more information about GPU offloading.
GPU assignement
"Manually assigning tasks to different GPUs is currently not supported, which means that number of GPU processes spawned will be equal to number of MPI ranks requested in slurm, and they will be evenly spread across GPUs."