NAMD
Description¶
NAMD is molecular dynamics simulation software using the Charm++ parallel programming model. It is noted for its parallel efficiency and is often used to simulate large systems (millions of atoms), develpoed by the collaboration of the Theoretical and Computational Biophysics Group (TCB) and the Parallel Programming Laboratory (PPL) at the University of Illinois at Urbana–Champaign.
Versions¶
Following versions of NAMD are currently available:
- Runtime dependencies:
You can load selected version (also with runtime dependencies) as a one module by following commands:
module load NAMD/2.14-foss-2021a-CUDA-11.3.1
User guide¶
You can find the software documentation and user guide on the official NAMD website and/or NAMD's user guide.
Benchmarks¶
In order to better understand how NAMD utilises the available hardware on Devana and how to get good performance we can examine the effect on benchmark performance of the choice of the number of MPI ranks per node and OpenMP thread.
Following command has been used to run the benchmarks:
mpiexec -np $ntmpi charmrun +p $ntomp +isomalloc_sync +setcpuaffinity namd2 +devices $gpu $trajectory
More information about these benchmarks systems, as well as downloadable input files, can be found here.
Info
"Single-node benchmarks have been run on local /work/ storage native to each compute node, which are generally faster than shared storage hosting /home/ and /scratch/ directories."
Benchmarks have been made on following systems:
Following file was used as input file for this benchmark:
NAMD Input file
#############################################################
## ADJUSTABLE PARAMETERS ##
#############################################################
structure stmv.psf
coordinates stmv.pdb
#############################################################
## SIMULATION PARAMETERS ##
#############################################################
# Input
paraTypeCharmm on
parameters par_all27_prot_na.inp
temperature 298
# Force-Field Parameters
exclude scaled1-4
1-4scaling 1.0
cutoff 12.
switching on
switchdist 10.
pairlistdist 13.5
# Integrator Parameters
timestep 1.0
nonbondedFreq 1
fullElectFrequency 4
stepspercycle 20
# Constant Temperature Control
langevin on ;# do langevin dynamics
langevinDamping 5 ;# damping coefficient (gamma) of 5/ps
langevinTemp 298
langevinHydrogen off ;# don't couple langevin bath to hydrogens
# Constant Pressure Control (variable volume)
useGroupPressure yes ;# needed for rigidBonds
useFlexibleCell no
useConstantArea no
langevinPiston on
langevinPistonTarget 1.01325 ;# in bar -> 1 atm
langevinPistonPeriod 100.
langevinPistonDecay 50.
langevinPistonTemp 298
cellBasisVector1 216.832 0. 0.
cellBasisVector2 0. 216.832 0.
cellBasisVector3 0. 0 216.832
cellOrigin 0. 0. 0.
PME on
PMEGridSizeX 216
PMEGridSizeY 216
PMEGridSizeZ 216
# Output
outputName /home/<username>/stmv-output
outputEnergies 20
outputTiming 20
numsteps 50000
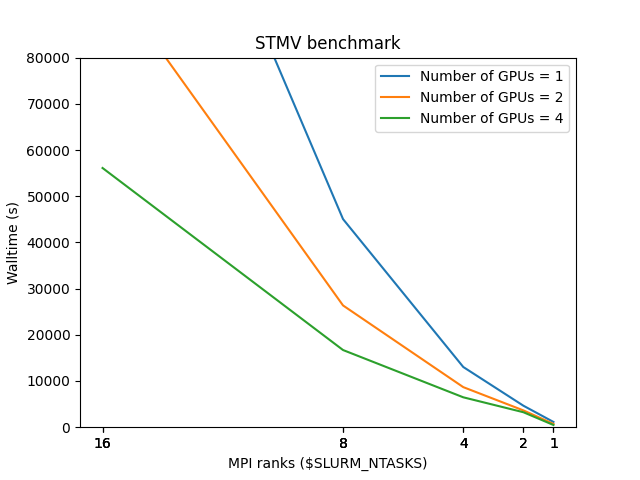
Numerical values for the benchmark are as follows:
| GPU/MPI | 1 | 2 | 4 | 8 | 16 |
|---|---|---|---|---|---|
| 1 | 1136 | 4652 | 13031 | 45043 | |
| 2 | 712 | 3635 | 8673 | 26355 | 99093 |
| 4 | 477 | 3241 | 6471 | 16703 | 56104 |
Info
Remaining combinations of MPI/OMP were not feasible due to memory limit on the GPUs.
Thus, users are adviced to run NAMD with single MPI rank and vary only number of cpus assigned to this rank. It is important to understand that if user requests one mpi rank (#SBATCH --ntasks-per-node=1) and let us say 32 cpus (#SBATCH --cpus-per-task=32) he/she will be charged 32 BUs. This should be taken into account when requesting number of gpus (#SBATCH --gres=gpu:2), as half the cpus on accelerated node allows usage of two accelerated processing units.
Generally speaking, the best combination of requested resources, with respect to BUs, are as follows:
| #SBATCH --ntasks-per-node= | #SBATCH --cpus-per-task= | #SBATCH --gres=gpu: |
|---|---|---|
| 1 | 16 | 1 |
| 2 | 32 | 2 |
| 3 | 48 | 3 |
| 4 | 64 | 4 |
Example run script¶
You can copy and modify this script to namd_run.sh and submit job to a compute node by command sbatch namd_run.sh.
GPU partition
NAMD is compiled only in GPU-aware version and must be run on gpu partition.
GPU IDs
Variable DEVICES in the following script refers to IDs of gpus, which range from 0 to 3. Users are highly adviced to use this keyword and to NOT pass the variable to the argument but rather use plain text. If one wishes to perform calculation on 1 GPU this part of command should look like +devices 0, for 2 GPUS use +devices 0,1, etc.
#!/bin/bash
#SBATCH -J "namd_job" # name of job in SLURM
#SBATCH --account=<project> # project number
#SBATCH --partition=gpu #
#SBATCH --nodes=1 # gpu allocations allows only single node
#SBATCH --ntasks-per-node= # number of mpi ranks per node
#SBATCH --cpus-per-task= # number of cpus per mpi rank
#SBATCH --time=hh:mm:ss # time limit for a job
#SBATCH -o stdout.%J.out # standard output
#SBATCH -e stderr.%J.out # error output
module load NAMD/2.14-foss-2021a-CUDA-11.3.1
# Modify according to specific needs
init_dir=`pwd`
work_dir=/work/$SLURM_JOB_ID
# Copy files over
input_files=""
output_files=""
# Move to working directory
cd $work_dir
cp $input_files $work_dir/.
# Start NAMD
mpiexec -np ${SLURM_NTASKS} charmrun +p ${SLURM_CPUS_PER_TAKS} +isomalloc_sync +setcpuaffinity namd2 +devices ${DEVICES} <input_file>
# Move files back
cp $output_files $init_dir/.