LAMMPS
Description¶
LAMMPS (Large-scale Atomic/Molecular Massively Parallel Simulator) is a classical molecular dynamics code with a focus on materials modeling. LAMMPS has potentials for solid-state materials (metals, semiconductors) and soft matter (biomolecules, polymers) and coarse-grained or mesoscopic systems. It can be used to model atoms or, more generically, as a parallel particle simulator at the atomic, meso, or continuum scale.
Versions¶
Following versions of LAMMPS package are currently available:
- Compiled with:
- PLUMED/2.7.2-foss-2021a
You can load selected version (also with runtime dependencies) as a one module by following command:
module load LAMMPS/23Jun2022-foss-2021a-kokkos
- Compiled with:
- PLUMED/2.8.2_MB-foss-2021a
You can load selected version (also with runtime dependencies) as a one module by following command:
module load LAMMPS/23Jun2022-foss-2021a-kokkos_MD
User guide¶
You can find the software documentation and user guide on the official LAMPPS website.
Optional Packages¶
Packages are groups of files that enable a specific set of features and extend LAMPPS functionality, such as force fields for molecular systems or rigid-body constraints, etc. These packages must be included as the part of the LAMMPS build proces. List and description of all packages can be found on offical LAMPPS website, section Package details.
Optional Packages
Current LAMMPS version includes only KOKKOS package. If you wish to have other packages included send a request to our administration support team.
Benchmarking LAMMPS¶
In order to better understand how LAMMPS utilises the available hardware on Devana and how to get good performance we can examine the effect on benchmark performance of the choice of the number of MPI ranks per node.
Following command has been used to run the benchmarks:
mpiexec -np $SLURM_NTASKS lmp -in ${benchmark}.inp > ${benchmark}.out
Info
Single-node benchmarks have been run on local /work/ storage native to each compute node, which are generally faster than shared storage hosting /home/ and /scratch/ directories.
OPENMP package
Current version of LAMMPS is compiled without OpenMP package that provides optimized and multi-threaded versions of many pair styles, nearly all bonded styles (bond, angle, dihedral, improper), several Kspace styles, and a few fix styles. Thus, the best performance can be achieved with maximum number of MPI ranks each running on single OMP thread, export OMP_NUM_THREADS=1.
Benchmarks have been made on following systems:
- 32,000 atoms for 100000 timesteps
- reduced density = 0.8442 (liquid)
- force cutoff = 2.5 sigma
- neighbor skin = 0.3 sigma
- neighbors/atom = 55 (within force cutoff)
- NVE time integration
| Single node Performance | Cross-node Performance |
|---|---|
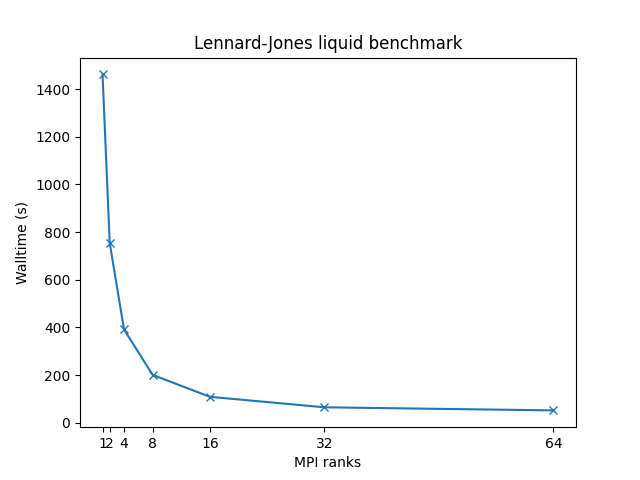 |
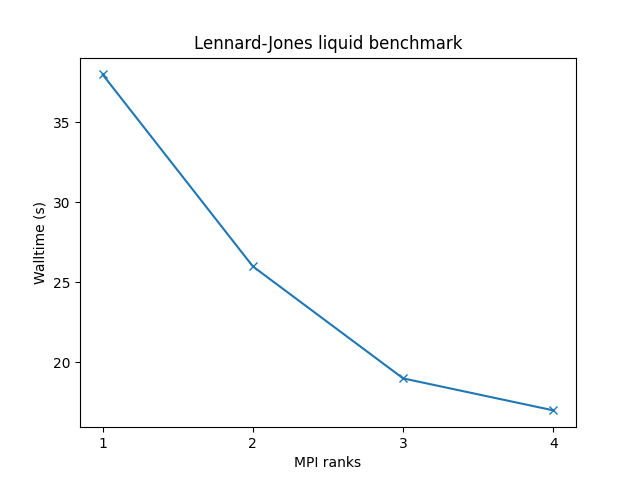 |
Bead-spring polymer melt with 100-mer chains and FENE bonds:
- 32,000 atoms for 100000 timesteps
- reduced density = 0.8442 (liquid)
- force cutoff = 2^(⅙) sigma
- neighbor skin = 0.4 sigma
- neighbors/atom = 5 (within force cutoff)
- NVE time integration
| Single node Performance | Cross-node Performance |
|---|---|
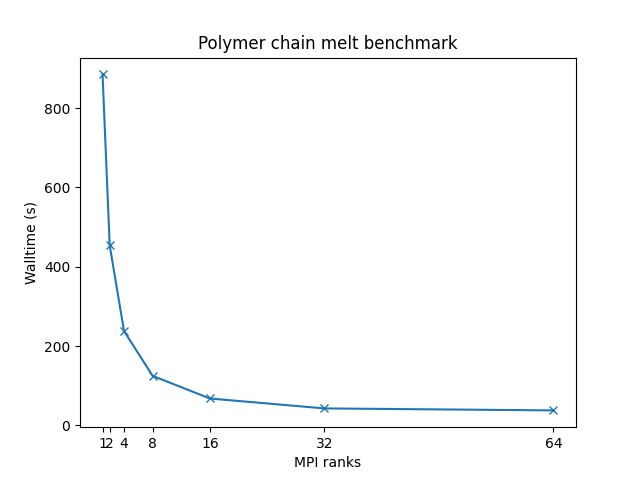 |
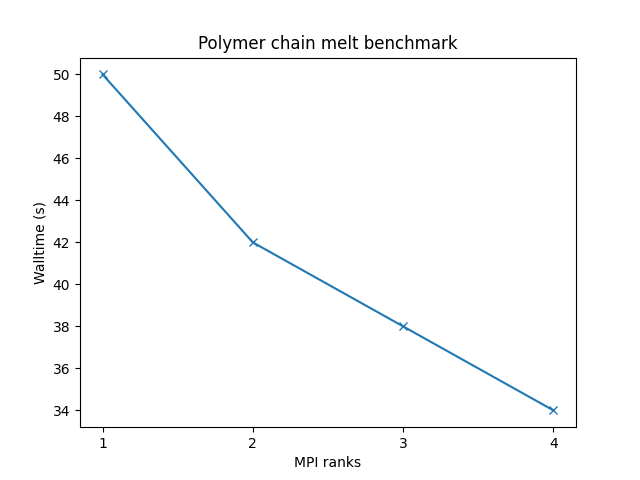 |
Cu metallic solid with embedded atom method (EAM) potential:
- 32,000 atoms for 100000 timesteps
- force cutoff = 4.95 sigma
- neighbor skin = 1.0 sigma
- neighbors/atom = 45 (within force cutoff)
- NVE time integration
| Single node Performance | Cross-node Performance |
|---|---|
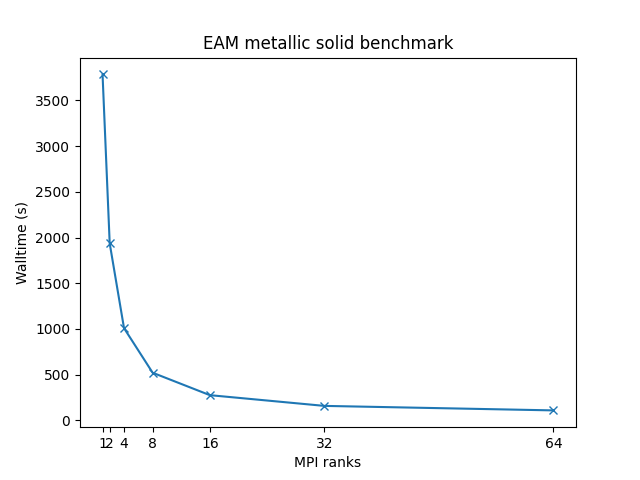 |
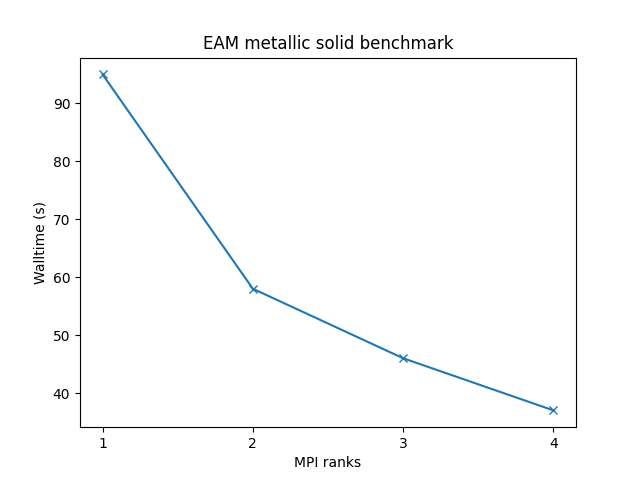 |
All-atom rhodopsin protein in solvated lipid bilayer with CHARMM force field, long-range Coulombics via PPPM (particle-particle particle mesh), SHAKE constraints:
- 32,000 atoms for 100000 timesteps
- LJ force cutoff = 10 A
- neighbor skin = 1.0 sigma
- neighbors/atom = 440 (within force cutoff)
- NPT time integration
| Single node Performance | Cross-node Performance |
|---|---|
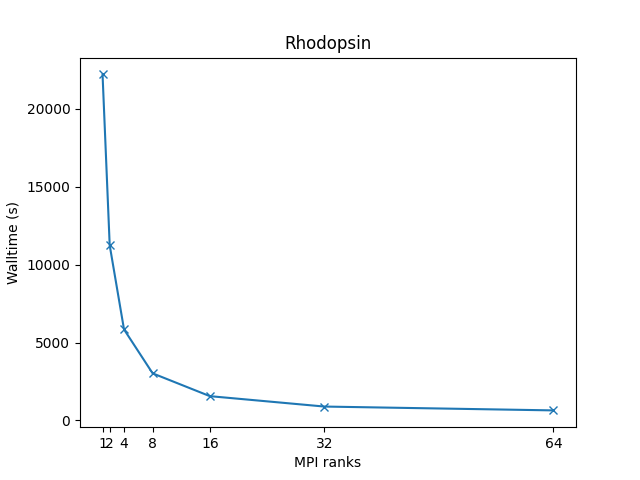 |
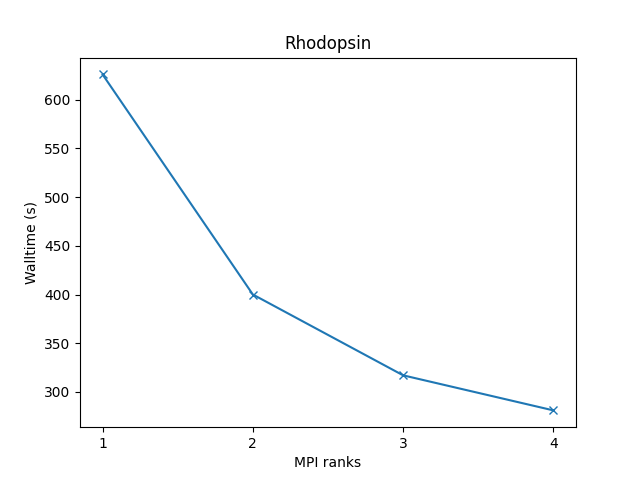 |
More information about these benchmarks, and many more examples, can be found on the official website designated to LAMMPS benchmarks.
Example run script¶
You can copy and modify this script to lammps_run.sh and submit job to a compute node by command sbatch lammps_run.sh.
#!/bin/bash
#SBATCH -J "LAMMPS_job" # name of job in SLURM
#SBATCH --account=<project> # project number
#SBATCH --partition= # select short, medium, long
#SBATCH --nodes= # number of nodes
#SBATCH --ntasks= # number of mpi ranks
#SBATCH --time=hh:mm:ss # time limit for a job
#SBATCH -o stdout.%J.out # standard output
#SBATCH -e stderr.%J.out # error output
module load LAMMPS/23Jun2022-foss-2021a-kokkos
# Modify according to specific needs
init_dir=`pwd`
work_dir=/scratch/$USER/LAMMPS/$SLURM_JOB_ID
# Copy files over
input_files=""
output_files=""
# Define input file
input=""
# Move to working directory
cd $work_dir
cp $input_files $work_dir/.
# Start LAMMPS
export OMP_NUM_THREADS=1
mpiexec -np ${SLURM_NTASKS} lmp -in ${input}.inp
# Move files back
cp $output_files $init_dir/.
GPU accelerated LAMMPS¶
Note
Currently, the GPU-aware version of LAMMPS is not available on Devana cluster.