NWChem
Description¶
NWChem aims to provide its users with computational chemistry tools that are scalable both in their ability to treat large scientific computational chemistry problems efficiently, and in their use of available parallel computing resources from high-performance parallel supercomputers to conventional workstation clusters. NWChem software can handle: biomolecules, nanostructures, and solid-state; from quantum to classical, and all combinations; Gaussian basis functions or plane-waves; scaling from one to thousands of processors; properties and relativity.
Versions¶
Following versions of NWChem are currently available:
- Runtime dependencies:
- intel/2022a
- GlobalArrays/5.8.1-intel-2022a
- Python/3.10.4-GCCcore-11.3.0
You can load selected version (also with runtime dependencies) as a one module by following command:
module load NWChem/7.0.2-intel-2022a
User guide¶
You can find the software documentation and user guide here.
Benchmarking¶
In order to better understand how NWChem utilises the available hardware on Devana and how to get good performance we can examine the effect on benchmark performance of the choice of the number of MPI ranks per node and OpenMP thread. For more information about these benchmarks systems see following NWChem benchmarks.
Following command has been used to run the MD simulation:
export OMP_NUM_THREADS=$nompt
mpiexec -np $ntmpi nwchem $input.inp > $output.out
Benchmarks have been made on the following systems:
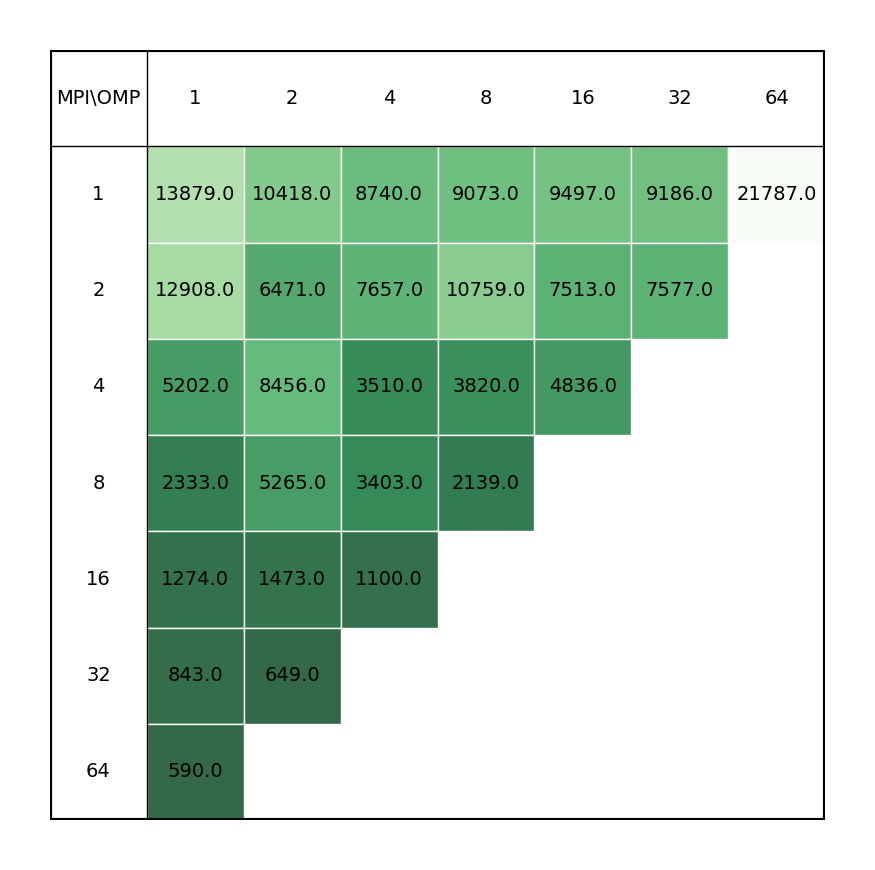
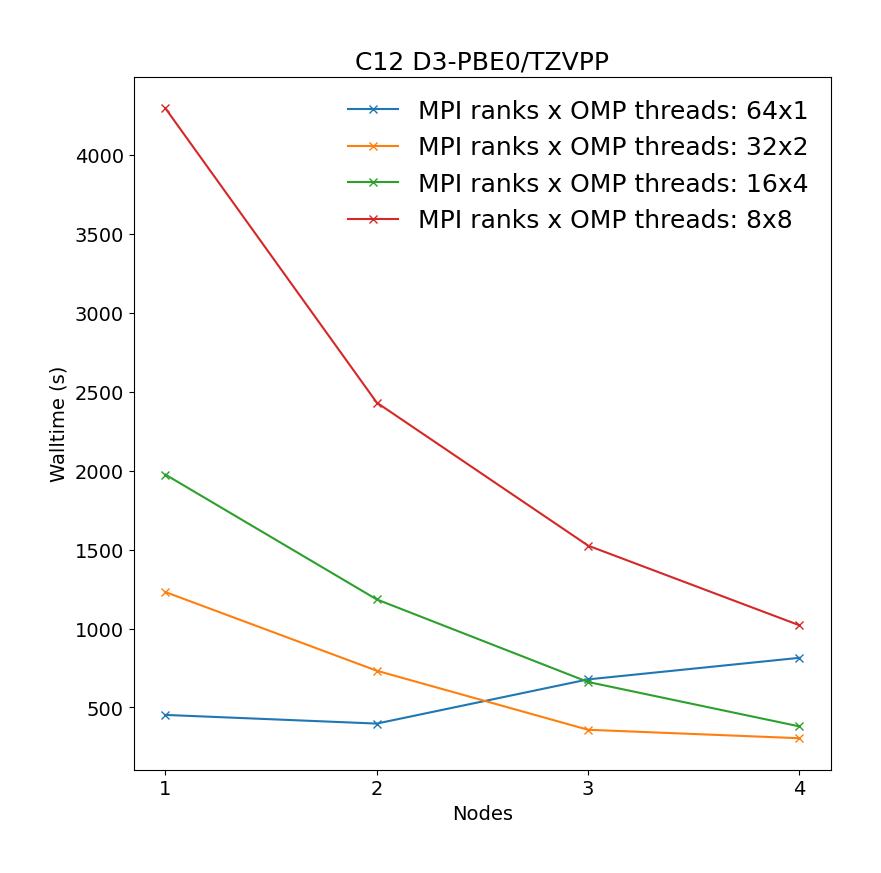
Geometry optimization of C12 benzene dimer at D3-PBE0/def2-TZVPP level of theory illustrating the most time-consuming part of the calculation, i.e. SCF calculations.
| Single node Performance | Cross-node Performance |
|---|---|
 |
 |
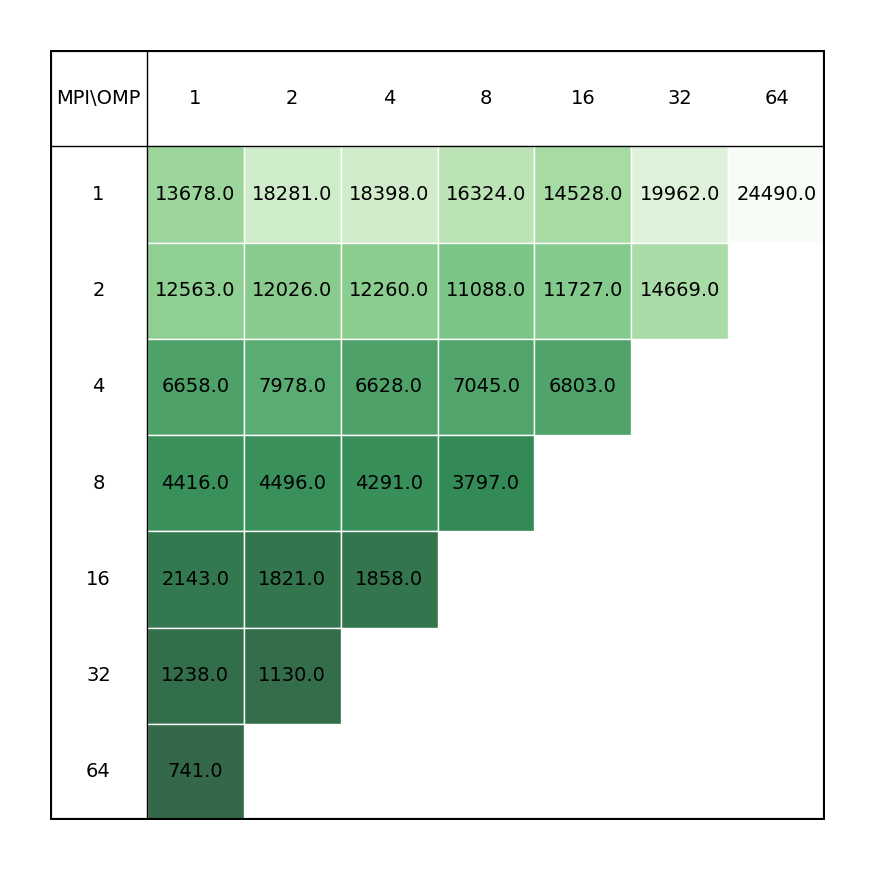
Geometry optimization of C36 fullerene at PBE0/6-31G* level of theory illustrating the most time-consuming part of the calculation, i.e. SCF calculations.
| Single node Performance | Cross-node Performance |
|---|---|
 |
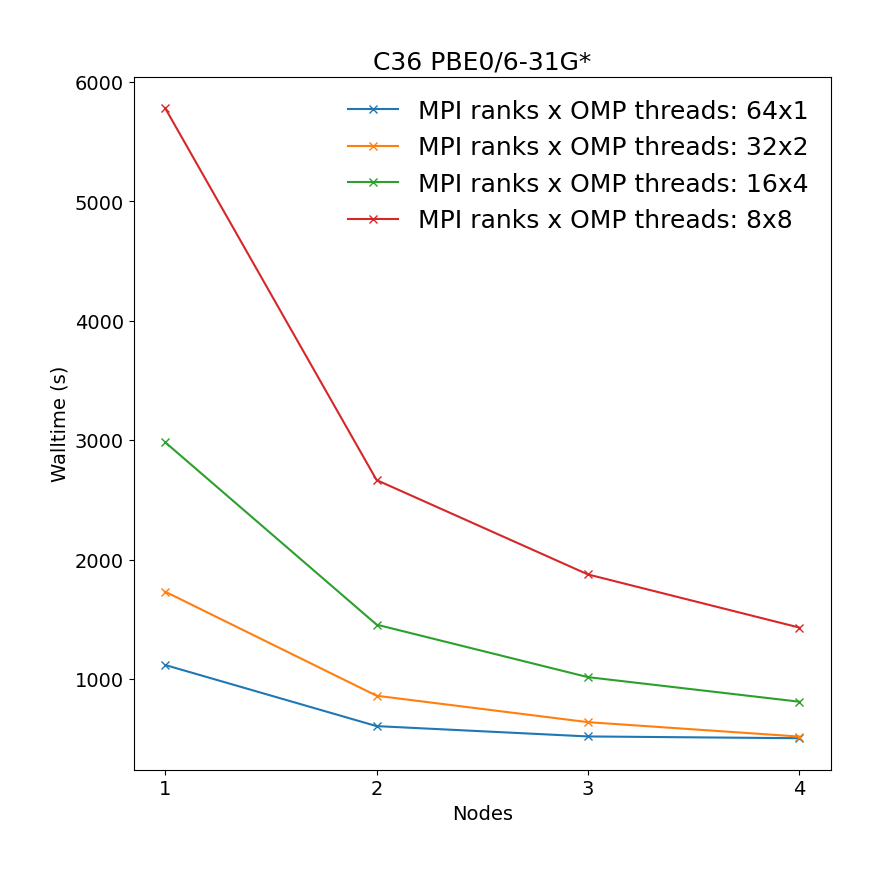 |
It is revealead that the best performance can be achieved by requesting maximum number of MPI ranks (ntasks), with respect to reserved portion of node/nodes, with each rank assigned one OMP thread.
Example run script¶
You can copy and modify this script to nwchem_run.sh and submit job to a compute node by command sbatch nwchem_run.sh.
#!/bin/bash
#SBATCH -J "nwchem_job" # name of job in SLURM
#SBATCH --account=<project> # project number
#SBATCH --partition= # selected partition (short, medium, long)
#SBATCH --nodes= # number of nodes
#SBATCH --ntasks= # number of mpi ranks (parallel run)
#SBATCH --time=hh:mm:ss # time limit for a job
#SBATCH -o stdout.%J.out # standard output
#SBATCH -e stderr.%J.out # error output
module load NWChem/7.0.2-intel-2022a
mkdir /scratch/$USER/NWChem/$SLURM_JOB_ID
export NWCHEM_SCRATCH_DIR=/scratch/$USER/NWChem/$SLURM_JOB_ID
export NWCHEM_PERMANENT_DIR=/scratch/$USER/NWChem/$SLURM_JOB_ID
INPUT=nwchem_input.inp
OUTPUT=nwchem_output.out
mpiexec -np $SLURM_NTASKS nwchem $INPUT > $OUTPUT # You can specify exact input and output name
GPU accelerated NWChem¶
Note
Currently, the GPU-aware version of NWChem is not available on Devana cluster.