Description¶
OpenMolcas is a quantum chemistry software package
Versions¶
Following versions of OpenMolcas package are currently available:
- Runtime dependencies:
- intel/2022a
- Python/3.10.4-GCCcore-11.3.0
- HDF5/1.12.2-iimpi-2022a
- GlobalArrays/5.8.1-intel-2022a
You can load selected version (also with runtime dependencies) as a one module by following command:
module load OpenMolcas/22.10-intel-2022a
User guide¶
You can find the software documentation and user guide on the official OpenMolcas website.
Benchmarks¶
OpenMolcas Parallelization
It is important to note that not all Molcas modules can benefit from parallel execution. This means that even though the calculation will get executed in parallel, all processes will perform the same serial calculation. This means that the benchmarks consisting of multiple modules do not gain the benefit of multiple MPI ranks throughout the full run. For list of parallelized Molcas modules see Parallelization efforts in Molcas manual.
Warning
The list of parallel modules is incomplete, as illustrated by COOH dimer benchmark with parallel execution of CHCC module, and the user is adviced to test the availability of parallel calculation on smaller systems, and/or inspect the specific modules in the OpenMolcas manual for any information about parallel calculations.
In order to better understand how OpenMolcas utilises the available hardware on Devana and how to get good performance we can examine the effect on benchmark performance of the choice of the number of MPI ranks per node and OpenMP thread.
Following command has been used to run the benchmarks:
export MOLCAS_WORKDIR=$scratch
export MOLCAS_MEM=$mem
pymolcas $molecule.inp -np $ntmpi -nt $ntomp -o $molecule.out
echo "scale=0; ($SLURM_MEM_PER_NODE / ( 64 * $ntomp )" | bc. Temporary working MOLCAS_WORKDIR directory was set to /scratch.
Benchmarks have been made on following systems:
Following file was used as input file for this benchmark:
OpenMolcas Input file
&SEWARD
Title= C2Au2
Symmetry = X Y Z
Basis set
C.ano-rcc-vtzp
C1 0.000000 0.000000 0.614833 Angstrom
End of basis
Basis set
Au.ano-rcc-vtzp
AU1 0.000000 0.000000 2.475008 Angstrom
End of basis
End of input
********************************************
&SCF
Title
C2Au2
Occupied
20 10 10 4 19 9 9 4
Iterations
70
Prorbitals
2 1.d+10
End of input
*****************************************
&RASSCF &END
Title
C2Au2
Symmetry
1
Spin
1
nActEl
2 0 0
Inactive
20 9 10 4 19 9 9 4
Ras2
0 1 0 0 0 0 0 0
Lumorb
ITERation
200 50
CIMX
200
PROR
100 0
THRS
1.0d-09 1.0d-05 1.0d-05
OutOrbitals
Canonical
End of input
****************************************
&MOTRA &END
JOBIph
Frozen
15 7 7 3 15 7 7 3
End of Input
****************************************
&CCSDT &End
Title
C2Au2 CC
CCT
ADAPtations
1
Denominators
2
T3DEnominators
0
TRIPles
3
Extrapolation
6,4
End of Input"
Note that only SEWARD, SCF, RASSCF modules are effectively parallelized in Molcas and not the coupled-cluster part of the calculation.
Following file was used as input file for this benchmark:
OpenMolcas Input file
&SEWARD
Title=COOH_dim
Cholesky
ChoInput
Thrc
0.00000001
EndChoInput
Basis set
C.cc-pvtz
C1 -1.888896 -0.179692 0.000000 Angstrom
C2 1.888896 0.179692 0.000000 Angstrom
End of basis
Basis set
O.cc-pvtz
O1 -1.493280 1.073689 0.000000 Angstrom
O2 -1.170435 -1.166590 0.000000 Angstrom
O3 1.493280 -1.073689 0.000000 Angstrom
O4 1.170435 1.166590 0.000000 Angstrom
End of basis
Basis set
H.cc-pvtz
H1 2.979488 0.258829 0.000000 Angstrom
H2 0.498833 -1.107195 0.000000 Angstrom
H3 -2.979488 -0.258829 0.000000 Angstrom
H4 -0.498833 1.107195 0.000000 Angstrom
End of basis
End of input
****************************************
&SCF
Title
COOH_dim
Occupied
24
Iterations
70
Prorbitals
2 1.d+10
End of input
****************************************
&CHCC &END
Title
CC part
Frozen
6
THRE
1.0d-08
End of input
****************************************
&CHT3 &END
Title
CC+T3 part
Frozen
6
End of input
Note that according to User Guide only SEWARD & SCF modules are effectively parallelized in Molcas the rest of calculation unable to profit from the parallel implementation. This was found to be innacurate during the benchmark, as even CHCC module was able to benefit from parallel calculation.
| C2Au2 aurocarbon | COOH dimer |
|---|---|
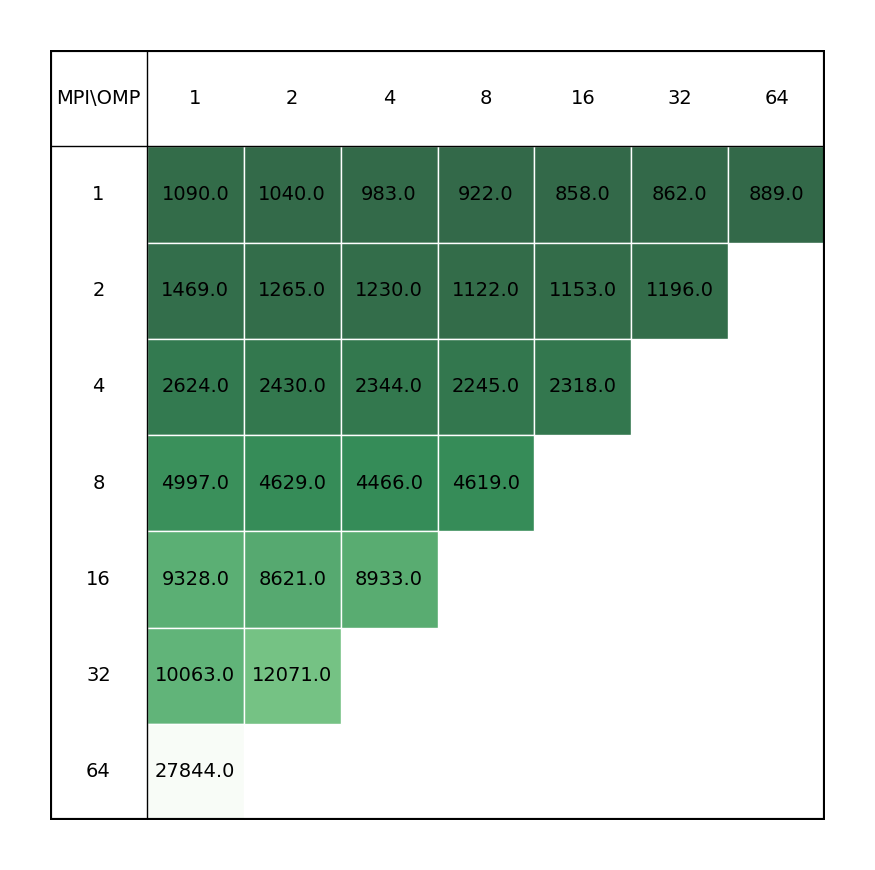 |
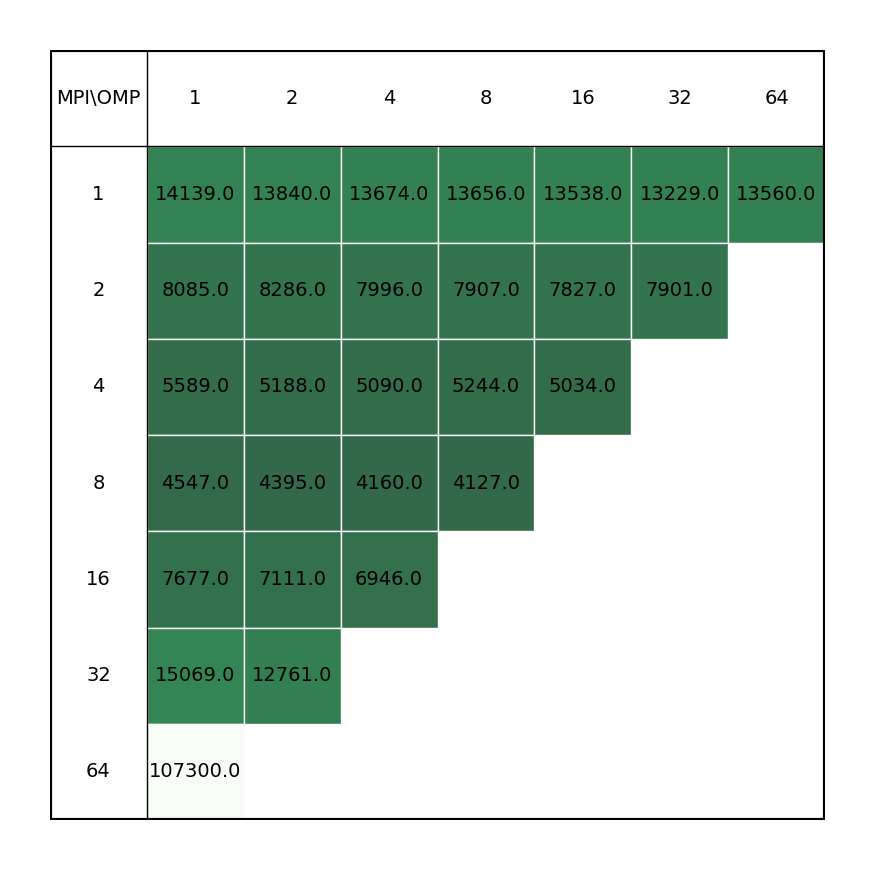 |
Users are not adviced to run OpenMolcas in parallel, unless their calculation is solely based on the modules that can benefit from parallelization scheme, or the non-parallelized part is inconsequential when compared to parallel modules. Inclusion of MPI parallelization to calculations containing significant part of non-paralallelized modules leads to a loss of performance, possible due to lesser available memory for each process (even for non-parallelized modules), which overshadows the gains from the parallelized parts of the calculation. The improved performance can be achieved by increasing the omp threads for the process, keyword -nt in the OpenMolcas command line.
Example run script¶
You can copy and modify this script to openmolcas_run.sh and submit job to a compute node by command sbatch openmolcas_run.sh.
#!/bin/bash
#SBATCH -J "openmolcas_job" # name of job in SLURM
#SBATCH --partition=ncpu # selected partition, short, medium or long
#SBATCH --nodes= # number of used nodes
#SBATCH --ntasks= # number of mpi tasks (parallel run)
#SBATCH --cpus-per-task= # number of mpi tasks (parallel run)
#SBATCH --time=72:00:00 # time limit for a job
#SBATCH -o stdout.%J.out # standard output
#SBATCH -e stderr.%J.out # error output
# Load modules
module load OpenMolcas/22.10-intel-2022a
# Create working directory for OpenMolcas
SCRATCH=/scratch/$SLURM_JOB_ACCOUNT/MOLCAS/$SLURM_JOB_ID
mkdir -p $SCRATCH
# Define memory per MPI rank
MEM= # One MPI rank on a single core corresponds to maximum of 3.9GB
# Export variables to OpenMolcas
export MOLCAS_WORKDIR=$SCRATCH
export MOLCAS_MEM=$MEM
INPUT=molcas_input.inp
OUTPUT=molcas_output.out
pymolcas $INPUT -np $SLURM_NTASKS -nt $SLURM_CPUS_PER_TASK -o $OUTPUT